| Research articles and book chapters | Popular science content | Opinion and policy | Patents |
Articles and book chapters
|
|
Engineering biology and the positive regulatory pathway in Brazil; |
|
|
A roadmap towards the synthesis of Life; |
|
|
Quencher-Free Fluorescence Monitoring of G-Quadruplex Folding; |
|
|
Confronting risks of mirror life; |
|
|
Technical Report on Mirror Bacteria: Feasibility and Risks; |
|
|
Engineering Biology for Space Health: An Innovative Research Roadmap; |
|
|
Laws of thought in living cells; |
|
|
Alternate conformational trajectories in ribosome translocation; |
|
|
preprint Nonenzymatic, prebiotic aminoacylation couples chirality of RNA and protein; |
|
|
Emergent ribozyme behaviors in oxychlorine brines indicate a unique niche for molecular evolution on Mars; |
|
|
Sequential gentle hydration increases encapsulation in model protocells; |
|
|
PACRAT: Pathogen detection with aptamer-detected cascaded recombinase polymerase amplification-in vitro transcription; |
|
|
Building Synthetic Cells─From the Technology Infrastructure to Cellular Entities; |
|
|
preprint Quencher-free fluorescence monitoring of G-Quadruplex folding; |
|
|
Preparing for the Future of Precision Medicine: Synthetic Cell Drug Regulation; |
|
|
Controlled exchange of protein and nucleic acid signals from and between synthetic minimal cells; |
|
|
Cell-Free Expressed Membraneless Organelles Inhibit Translation in Synthetic Cells; |
|
|
preprint High yield, low magnesium flexizyme reactions in a water-ice eutectic phase; |
|
|
Present and future of synthetic cell development; |
|
|
Trumpet is an operating system for simple and robust cell-free biocomputing; |
|
|
Encyclopedia of Astrobiology; |
|
|
Cell-Free Expression System Derived from a Near-Minimal Synthetic Bacterium; |
|
|
A gene expression control technology for cell-free systems and synthetic cells via targeted gene silencing and transfection; |
|
|
New Aequorea fluorescent proteins for cell‐free bioengineering; |
|
|
Parasites, infections and inoculation in synthetic minimal cells; |
|
|
T7Max transcription system |
|
|
A ubiquitous amino acid source for prokaryotic and eukaryotic cell-free transcription-translation systems |
|
|
Engineering Ribosomes to Alleviate Abiotic Stress in Plants; |
|
|
Expanding luciferase reporter systems for cell-free protein expression; |
|
|
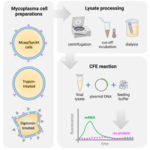
Traditional protocols and optimization methods lead to absent expression in mycoplasma cell-free gene expression platform |
|
|
Akaby - cell-free protein expression system for linear templates |
|
|
Diffusion control in biochemical specificity; |
|
|
Switchable DNA-based Peroxidases Controlled by a Chaotropic Ion; |
|
|
Making Security Viral: Shifting Engineering Biology Culture and Publishing; |
|
|
Liposome Preparation by 3D-Printed Microcapillary-Based Apparatus; |
Nathaniel J. Gaut, Jose Gomez-Garcia, Joseph M. Heili, Brock Cash, Qiyuan Han, Aaron E. Engelhart, Katarzyna P. Adamala
ACS Syn Bio, 2022, https://doi.org/10.1021/acssynbio.1c00519
|
|
Build-a-Cell: Engineering a Synthetic Cell Community |
|
|
Synthetic Cells in Biomedical Applications |
|
|
Building a community to engineer synthetic cells and organelles from the bottom-up |
|
|
Toward synthetic life: Biomimetic synthetic cell communication |
|
|
Reconstituting Natural Cell Elements in Synthetic Cells |
|
|
Spatial multiplexing of fluorescent reporters for dynamic imaging of signal transduction networks |
|
|
Rapid deployment of smartphone‐based augmented reality tools for field and online education in structural biology |
|
|
Highly specific, multiplexed isothermal pathogen detection with fluorescent aptamer readout |
|
|
Toward artificial photosynthesis Nathaniel J Gaut and Katarzyna P Adamala Science Vol. 368, Issue 6491, pp. 587-588 journal link |
|
|
Connectome of synthetic cells: Comment on “What would a synthetic connectome look like?” by Ithai Rabinowitch; |
|
|
Enabling community-based metrology for wood-degrading fungi; |
Tanner G. Hoog, Matthew R. Pawlak, Lauren M. Aufdembrink, Benjamin R. Bachan, Matthew B. Galles, Nicholas B. Bense, Katarzyna P. Adamala, Aaron E. Engelhart
https://doi.org/10.1101/784561
|
|
Biology on sample size of more than one |
Sivan Kaminski Strauss, (...) Katarzyna P. Adamala, (...), Orna Dahan, Yitzhak Pilpel;
Plos Biology, doi.org/10.1371/journal.pbio.3000182;
local copy
publisher website link
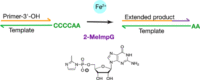
Lin Jin, Aaron E. Engelhart, Weicheng Zhang, Katarzyna Adamala, Jack W. Szostak
J. Am. Chem. Soc. 2018, 140, 44, 15016-15021, doi.org/10.1021/jacs.8b09617
Joseph M. Heili, Jose Gomez-Garcia, Nathaniel J. Gaut, Brock W. Cash, Lauren M. Aufdembrink, Brent A. Heffron, Joshua D. Shirley, Erin E. Carlson, Katarzyna P. Adamala, and Aaron E. Engelhart
J. Chem. Educ.,, 2018, 95 (10), pp 1867–1871; DOI: 10.1021/acs.jchemed.7b00735
local copy pdf publisher website

|
|
What Is the Role of Circuit Design in the Advancement of Synthetic Biology? |
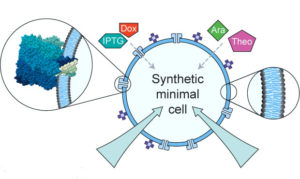
Engineering genetic circuit interactions within and between synthetic minimal cells;
Katarzyna P. Adamala*, Daniel A. Martin-Alarcon*, Katriona R. Guthrie-Honea, Edward S. Boyden;
Nature Chemistry, 9, 431-439, 2017, doi:10.1038/nchem.2644; *equal contribution
publisher website link
local copy pdf

Astrobiology Primer 2.0.;
Domagal-Goldman and Wright et al. (includes chapter editor K.P.Adamala)
Astrobiology, 2016, 16(8):561-653. doi:10.1089/ast.2015.1460
publisher website link
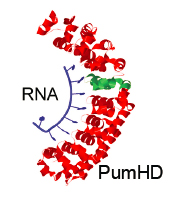
Programmable RNA-binding protein composed of repeats of a single modular unit;
Katarzyna P. Adamala*, Daniel A. Martin-Alarcon*, and Edward S. Boyden;
PNAS, 2016, 10.1073/pnas.1519368113; *equal contribution
local copy pdf
publisher website link
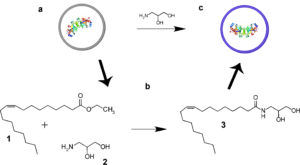
Collaboration between primitive cell membranes and soluble catalysts;
K. Adamala*, A. E. Engelhart* and J. W. Szostak;
Nature Communications, 2016, doi:10.1038/ncomms11041; *equal contribution
local copy pdf
publisher website link
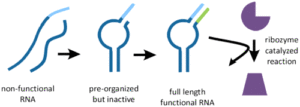
Generation of Functional RNAs from Inactive Oligonucleotide Complexes by Non-enzymatic Primer Extension;
K. Adamala*, A. E. Engelhart* and J. W. Szostak;
J. Am. Chem. Soc., 2015, 137 (1), pp 483 - 489, DOI: 10.1021/ja511564d; *equal contribution
local copy pdf
publisher website link
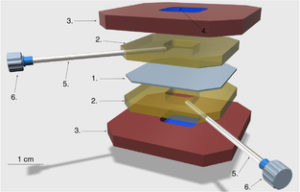
Construction of a Liposome Dialyzer for preparation of high-value, small-volume liposome formulations;
K. Adamala*, A. E. Engelhart*, N. Kamat, L. Jin and J. W. Szostak;
Nature Protocols, 2015, 10(6), pp 927-938; *equal contribution
local copy pdf
publisher website link
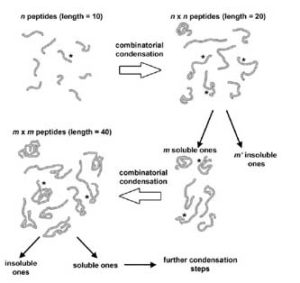
Open questions in origin of life: experimental studies on the origin of nucleic acids and proteins with specific and functional sequences by a chemical synthetic biology approach;
Adamala K, Anella F, Wieczorek R, Stano P, Chiarabelli C, Luisi PL;
Comput Struct Biotechnol J. 2014;9:e201402004
local copy pdf
publisher website link
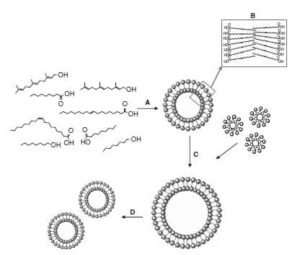
Experimental systems to explore life origin: perspectives for understanding primitive mechanisms of cell division;
K. Adamala, P.L. Luisi;
Results Probl. Cell. Differ. 53: 1-9.
local copy pdf
publisher website link

Non-enzymatic template-directed RNA synthesis inside model protocells;
K. Adamala and J.W. Szostak,
Science 342 (2013) 1098 - 1100;
local copy pdf
publisher website link
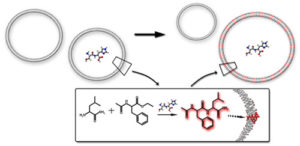
Competition between model protocells driven by an encapsulated catalyst;
K. Adamala and J.W. Szostak,
Nature Chemistry 5 (2013) 495 - 501;
local copy pdf
publisher website link
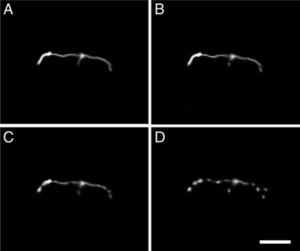
Photochemically driven redox chemistry induces protocell membrane pearling and division; T. F. Zhu, K. Adamala, N. Zhang, J. W. Szostak;
PNAS, 109 (2012) 9828–9832;
local copy pdf
publisher website link
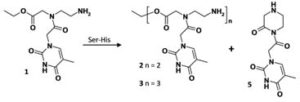
Ser-His catalyses the formation of peptides and PNAs;
M. Gorlero, R. Wieczorek, K. Adamala, A. Giorgi, M.E. Schinina, P. Stano, P.L. Luisi;
FEBS Letters, 583 (2009) 153 - 156;
local copy pdf
publisher website link
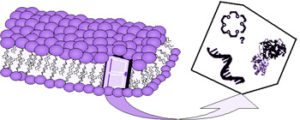
How to close the door leaving it open? On the origin of membrane transport system;
K. Adamala International Journal of Astrobiology, 7-1 (2008) 74;
Conference proceedings (selected)
Cleaves, Henderson James; Goldman, Aaron David; Jia, Tony Z; Keating, Christine; Adamala, Kate; Ditzler, Mark A; Maurer, Sarah; 'Major transitions in organismal individuality: From compartments, to protocells, to cells 2019 Astrobiology Science Conference, 2019
Engelhart, Aaron; Gaut, Nathaniel; Pawlak, Matt; Bachan, Benjamin; Adamala, Kate; 'The RNA World on Mars' 2019 Astrobiology Science Conference, 2019
Heili, Joseph; Gaut, Nathaniel; Han, Qiyuan; Gomez-Garcia, Jose; Szostak, JW; Adamala, KP; Engelhart, AE; 'Functional Interactions Between Early Biopolymers and Primitive Cells' LPICo, 2017 1967 4207
Tourlomousis, F; Karydis, T; Johnson, S; Adamala, K; Chang, RC; Mershin, A; 'Digital Additive Biomanufacturing of 3D Cell-Synell Co-Cultures' Tissue Engineering Part A, 2017 23 S52-S52
Gaut, Nathaniel; Heili, Joseph; Gomez-Garcia, Jose; Engelhart, Aaron; Adamala, Kate; 'Pre-LUCA cells: life but not alive' LPICo, 2017 1967 4200
Martin-Alarcon, Daniel; Adamala, Katarzyna; Guthrie-Honea, Katriona; 'A modular protein toolbox for RNA targeting' Synthetic Biology: Engineering, Evolution, and Design Conference 2015, SEED 2015, 2015 508-520
Adamala, K; Gochna, M; Gorski, M; Kowalska, I; 'SOWA-Polish National Astrobiology Student's Conference' International Journal of Astrobiology, 2008 7 1 90-90
Adamala, Katarzyna; Luisi, Pier Luigi; 'Experimental systems to explore life origin: perspectives for understanding primitive mechanisms of cell division' Cell Cycle in Development, 2011 43839
Patents
Engineering genetic circuit interactions within and between synthetic minimal cells and use therefor
ES Boyden, KP Adamala, DA Martin‐Alarcon
US Patent App. 62/294,586, 2016
Spatial multiplexing for multisignal cellular imaging
G Xu, E Boyden, KD Piatkevich, K Adamala
US Patent App. 15/099,232, 2016
outside link
Pumilio Domain-based Modular Protein Architecture for RNA Binding
ES Boyden, KP Adamala, DA Martin-Alarcon
US Patent App. 14/995,169
outside link
Popular science content by us and about us
Creating a New Tree of Life in the Lab: Threats and Opportunities
Thinking Beyond presents: Creating a New Tree of Life in the Lab: Threats and Opportunities
The “Thinking Beyond” webinar series features renowned physicists Paul Davies, Sara Walker and Maulik Parikh from the Beyond Center for Fundamental Concepts in Science at Arizona State University and addresses big questions/topics in science.
Life, but not alive
Life, But Not Alive - synthetic cell engineering at Concordia 4th Space
The 4TH SPACE Concordia University organized public seminar on synthetic cell engineering.
The Conversation on mirror life
Mirror life is a scientific fantasy leading to a dangerous reality
![]()
https://theconversation.com/mirror-life-is-a-scientific-fantasy-leading-to-a-dangerous-reality-a-synthetic-biologist-explains-how-mirror-bacteria-could-conquer-life-on-earth-245842
Mirror cell risks explained
Mirror Bacteria Research Poses Significant Risks, Dozens of Scientists Warn

https://www.the-scientist.com/mirror-bacteria-research-poses-significant-risks-dozens-of-scientists-warn-72419
Will the origin of life ever be uncovered? | Kate Adamala, Addy Pross and Chrisantha Fernando
Scientists disagree on the origin of life
Will the origin of life ever be uncovered? | Kate Adamala, Addy Pross and Chrisantha Fernando
Discussion on the origin(s) of life with Kate Adamala, Addy Pross and Chrisantha Fernando
Quanta podcast
Quanta Magazine podcast, "Even Synthetic Life Forms With a Tiny Genome Can Evolve".
By watching “minimal” cells regain the fitness they lost, researchers are testing whether a genome can be too simple to evolve.

https://podcasts.apple.com/us/podcast/even-synthetic-life-forms-with-a-tiny-genome-can-evolve/id1021340531?i=1000641216723/
The Art and Science of Synthetic Biology
Kate talked about engineering cells from scratch for biotechnology applications.
the-scientist.com/podcasts/the-art-and-science-of-synthetic-biology-71525
podbean link: thescientistspeaks.podbean.com/e/the-art-and-science-of-synthetic-biology

Building life in the lab
Kate talked about engineering synthetic cells as part of SynBYSS seminar series. Recording on youtube.
On evolution of minimal cells
Excellent summary of work on the evolution of minimal living cells.
Trumpet in the news
Kate and Judee talk about Trumpet biocomputing platform and practical applications for medicine.

Trumpet biocomputing
Our Trumpet platform summarized with emphasis on biomedical applications.

Building life in the lab - Reason with Science
Kate talked about building life in the lab on the Reason with Science channel. Recording on youtube or podcast website.
Neha Anwar podcast on synthetic cells
Kate was guest of the Neha Anwar podcast, talking about biomedical and other practical applications of synthetic cell engineering.
Synthetic cell engineering podcast.
YoloTalks: Creating artificial cells
Kate was guest of the YoloTalks podcast, talking about artificial cells and space exploration. Youtube: Creating artificial cells, and Spotify version.
Life as we don't know it
Our lab was featured in CBS Connect article "Life as we don't know it".
webinar Building a Synthetic Biology Platform for Drug Delivery
Kate participates in webinar "Building a Synthetic Biology Platform for Drug Delivery" organized by The Scientist.
![]()
Synthetic cell pathway to FDA
![]()
https://www.statnews.com/2022/07/26/fda-develop-framework-evaluate-synthetic-cells/
Building Artificial Cells with Kate Adamala | Late Night Conference
Kate Adamala wants to make life from scratch! As Professor at University of Minnesota working on the origins of life and building a synthetic cell, Kate’s work touches on astrobiology, synthetic cell engineering, and biocomputing. Kate and her team create tiny bioreactors. These have applications in synthetic biology, drug development and biosensing. Building Artificial Cells with Kate Adamala | Late Night Conference with Wilhelm Huck 2x05.
The life of our Last Universal Common Ancestor
Synthetic biology allows us to build minimal cell-like systems, to investigate origin of life and build tools for space exploration.
The life of our Last Universal Common Ancestor | LAS 2021.
Artificial cells’ valuable niche in medicine
Advanced Science News piece:
Artificial cells with specialized internal chemistries could revolutionize how we approach precision medicine.

https://www.advancedsciencenews.com/artificial-cells-valuable-niche-in-medicine/
webinar on synthetic cell engineering
Kate participates in The Dissenter webinar "Synthetic Cells, Cell Evolution, the Origin of Life, and Astrobiology".
EBRC podcast on safety of synthetic cells
Kate was a guest of EBRC In Translation podcast, discussing science, safety and security of synthetic cell engineering ebrcintranslation.buzzsprout.com/1581817/9465178-9-building-cells-and-synbio-in-space-w-kate-adamala

Synthetic cells for drug delivery
CRS News published our text about synthetic cells in drug delivery applications.
Synthetic cells: happy middle between liposome drug delivery and engineered natural cells

iBiology synthetic cells
Kate's iBiology talks on synthetic cells have been published.
Synthetic Cells: Building Life to Understand It

Part 1: Synthetic Cells: Building Life to Understand It

Part 2: Synthetic Cells: Approach and Applications
webinar Synthetic Cells · Artificial Life · Brain-Computer Interfaces · Space Exploration
Kate participates in webinar Synthetic Cells · Artificial Life · Brain-Computer Interfaces · Space Exploration hosted by the Molecular Programming Interest Group.

Life but not alive - Origins Center
Webinars of the Origins Center, the Netherlands: "Life but not alive"
Life but not alive.
Space Explr podcast on building synthetic cells
Kate was a guest of Space Explr podcast, discussing Science And Engineering of Building Synthetic Cells https://www.youtube.com/watch?v=4fznl9wLtmw
Probable Meets Possible: When Life Gets Weird
Kate Adamala and Trinity Hamilton from the College of Biological Sciences at the University of Minnesota discuss how tiny life forms on the fringes may illuminate huge truths about life in the universe. What do microbes that live on glaciers and in snow on Earth tell us about the potential for life on Mars? What can a cell made from scratch in a lab reveal about how life may have originated here on Earth—and beyond? This research on extremophiles and synthetic cells provides a unique take on life that may shed important insights into space travel and extraterrestrial life.
Probable Meets Possible: When Life Gets Weird.
Community of Biotechnology talk
Kate gave a seminar for the Community of Biotechnology.
https://www.facebook.com/cbiotechnology/videos/680207569301004/
Conversations with Aanika E10: Creating Synthetic Cells with Dr. Kate Adamala
Aanika Biosciences presents conversation series on topics of cutting edge synthetic biology.
Conversations with Aanika E10: Creating Synthetic Cells with Dr. Kate Adamala.
Quanta podcast
Kate helped with Quanta Magazine Bacterial Complexity Revises Ideas About ‘Which Came First?’ podcast.
https://www.quantamagazine.org/bacterial-organelles-revise-ideas-about-which-came-first-20190612/
Lab Work Under Isolation
Opinion on lab operations in The Scientist
https://www.the-scientist.com/opinion-lab-work-under-isolation-67398
Synthetic cell talk online
Kate's Waterloo seminar is posted online. Primer on synthetic cell work in our lab and little bit of the overview of the field.
P3 Dystopia
Kate's episode on P3 Dystopia show is now live.
https://sverigesradio.se/artikel/7345117
Origins of Life: Chemical Comminalties - Early Metabolisms - An Introduction
ComplexityExplorer.org course 'Origins of Life
Origins of Life: Chemical Comminalties - Early Metabolisms - An Introduction by Kate Adamala.
Mindscape podcast
Kate's episode on Sean Carroll's Mindscape podcast is live.
https://www.preposterousuniverse.com/podcast/2019/07/22/56-kate-adamala-on-creating-synthetic-life/

Life, But Not Alive: Breakthrough Discuss 2019
Breakthrough Discuss 2019 was held at UC Berkeley April 11-12, 2019. The theme, “Migration of Life in the Universe,” brought together an all-star group of astronomers, biologists, chemists and other experts to discuss whether and how primitive life can move between planets and stars, and if that is how life began on Earth.
Kate Adamala | Life, But Not Alive: Breakthrough Discuss 2019 .
Artificial cells gain communication skills
Great editorial in Science intorducing new, exciting paper and highlighting interesting aspects of building synthetic cells.
How biologists are creating life-like cells from scratch
Excellen editorial in Nature, describing rationale of engineering synthetic life, and the most common approaches to various problems we're facing en route to a ynthetic cell. 
Local copy pdf
From chemicals to life: Scientists try to build cells from scratch
Interesting summary of the origins of Build-a-Cell initiative.
Genetic circuits in synthetic cells
Kate published commentary on using synthetic cells to prototype cell circuits in Cell Systems journal.
Personalized medicine
Kate's Medium post Why should we personalize medicine?
TEDx Talk on Synthetic life
Kate presented concept of building synthetic minimal cells, with its biotechnological, biomedical and basic science implications, in a TEDx talk "Life but not alive".
Op-eds and policy papers
The Conversation on mirror life
Mirror life is a scientific fantasy leading to a dangerous reality
![]()
https://theconversation.com/mirror-life-is-a-scientific-fantasy-leading-to-a-dangerous-reality-a-synthetic-biologist-explains-how-mirror-bacteria-could-conquer-life-on-earth-245842
Mirror cell risks explained
Mirror Bacteria Research Poses Significant Risks, Dozens of Scientists Warn

https://www.the-scientist.com/mirror-bacteria-research-poses-significant-risks-dozens-of-scientists-warn-72419
Synthetic cell pathway to FDA
![]()
https://www.statnews.com/2022/07/26/fda-develop-framework-evaluate-synthetic-cells/
Artificial cells’ valuable niche in medicine
Advanced Science News piece:
Artificial cells with specialized internal chemistries could revolutionize how we approach precision medicine.

https://www.advancedsciencenews.com/artificial-cells-valuable-niche-in-medicine/
Lab Work Under Isolation
Opinion on lab operations in The Scientist
https://www.the-scientist.com/opinion-lab-work-under-isolation-67398