News and updates
@KateAdamala
2025-04-02 23:48:17
Creating a New Tree of Life in the Lab: Threats and Opportunities
Thinking Beyond presents: Creating a New Tree of Life in the Lab: Threats and Opportunities
The “Thinking Beyond” webinar series features renowned physicists Paul Davies, Sara Walker and Maulik Parikh from the Beyond Center for Fundamental Concepts in Science at Arizona State University and addresses big questions/topics in science.
2025-03-22 21:23:06
new publication
|
|
Engineering biology and the positive regulatory pathway in Brazil; |
2025-03-20 23:34:58
Life, but not alive
Life, But Not Alive - synthetic cell engineering at Concordia 4th Space
The 4TH SPACE Concordia University organized public seminar on synthetic cell engineering.
2025-02-11 21:37:40
The Conversation on mirror life
Mirror life is a scientific fantasy leading to a dangerous reality
![]()
https://theconversation.com/mirror-life-is-a-scientific-fantasy-leading-to-a-dangerous-reality-a-synthetic-biologist-explains-how-mirror-bacteria-could-conquer-life-on-earth-245842
2025-02-06 00:17:48
new publication
|
|
A roadmap towards the synthesis of Life; |
2025-01-30 19:35:43
new publication
|
|
Quencher-Free Fluorescence Monitoring of G-Quadruplex Folding; |
2024-12-18 20:18:53
Mirror cell risks explained
Mirror Bacteria Research Poses Significant Risks, Dozens of Scientists Warn

https://www.the-scientist.com/mirror-bacteria-research-poses-significant-risks-dozens-of-scientists-warn-72419
2024-12-16 22:10:25
new publication
|
|
Confronting risks of mirror life; |
2024-12-03 21:47:26
new publication
|
|
Technical Report on Mirror Bacteria: Feasibility and Risks; |
2024-11-07 21:06:11
new publication
|
|
Engineering Biology for Space Health: An Innovative Research Roadmap; |
2024-09-10 20:32:58
new publication
|
|
Laws of thought in living cells; |
2024-08-14 02:45:21
new publication
|
|
Alternate conformational trajectories in ribosome translocation; |
2024-07-31 19:46:15
new publication
|
|
preprint Nonenzymatic, prebiotic aminoacylation couples chirality of RNA and protein; |
2024-05-20 17:03:08
new publication
|
|
Emergent ribozyme behaviors in oxychlorine brines indicate a unique niche for molecular evolution on Mars; |
2024-05-16 18:58:23
new publication
|
|
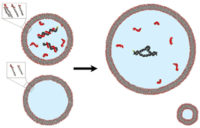
Sequential gentle hydration increases encapsulation in model protocells; |
2024-05-13 01:06:49
Will the origin of life ever be uncovered? | Kate Adamala, Addy Pross and Chrisantha Fernando
Scientists disagree on the origin of life
2024-04-24 14:11:08
new publication
|
|
PACRAT: Pathogen detection with aptamer-detected cascaded recombinase polymerase amplification-in vitro transcription; |
2024-03-26 23:58:21
new publication
|
|
Building Synthetic Cells─From the Technology Infrastructure to Cellular Entities; |
2024-03-08 00:38:32
Will the origin of life ever be uncovered? | Kate Adamala, Addy Pross and Chrisantha Fernando
Discussion on the origin(s) of life with Kate Adamala, Addy Pross and Chrisantha Fernando
2024-02-07 21:07:10
Quanta podcast
Quanta Magazine podcast, "Even Synthetic Life Forms With a Tiny Genome Can Evolve".
By watching “minimal” cells regain the fitness they lost, researchers are testing whether a genome can be too simple to evolve.

https://podcasts.apple.com/us/podcast/even-synthetic-life-forms-with-a-tiny-genome-can-evolve/id1021340531?i=1000641216723/
2024-02-04 02:41:49
new publication
|
|
preprint Quencher-free fluorescence monitoring of G-Quadruplex folding; |
2024-01-25 01:57:50
new publication
|
|
Preparing for the Future of Precision Medicine: Synthetic Cell Drug Regulation; |
2024-01-16 19:16:48
new publication
|
|
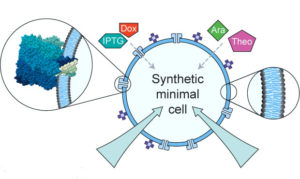
Controlled exchange of protein and nucleic acid signals from and between synthetic minimal cells; |
2024-01-14 01:25:02
new publication
|
|
Cell-Free Expressed Membraneless Organelles Inhibit Translation in Synthetic Cells; |
2023-12-18 23:49:38
new publication
|
|
preprint High yield, low magnesium flexizyme reactions in a water-ice eutectic phase; |
2023-12-16 00:19:44
new publication
|
|
Present and future of synthetic cell development; |
2023-11-29 22:32:03
The Art and Science of Synthetic Biology
Kate talked about engineering cells from scratch for biotechnology applications.
the-scientist.com/podcasts/the-art-and-science-of-synthetic-biology-71525
podbean link: thescientistspeaks.podbean.com/e/the-art-and-science-of-synthetic-biology

2023-10-29 23:43:26
new publication
|
|
Trumpet is an operating system for simple and robust cell-free biocomputing; |
2023-10-15 21:22:49
Building life in the lab
Kate talked about engineering synthetic cells as part of SynBYSS seminar series. Recording on youtube.
2023-08-10 21:21:56
On evolution of minimal cells
Excellent summary of work on the evolution of minimal living cells.
2023-08-10 20:16:08
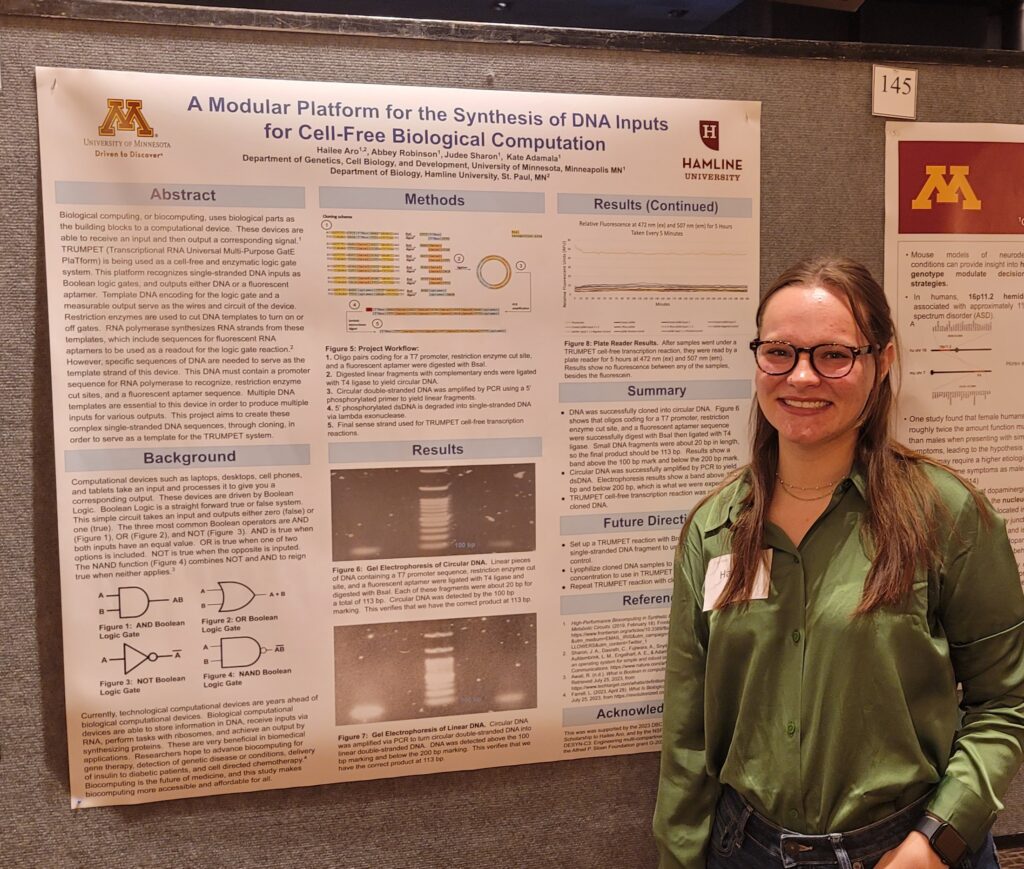
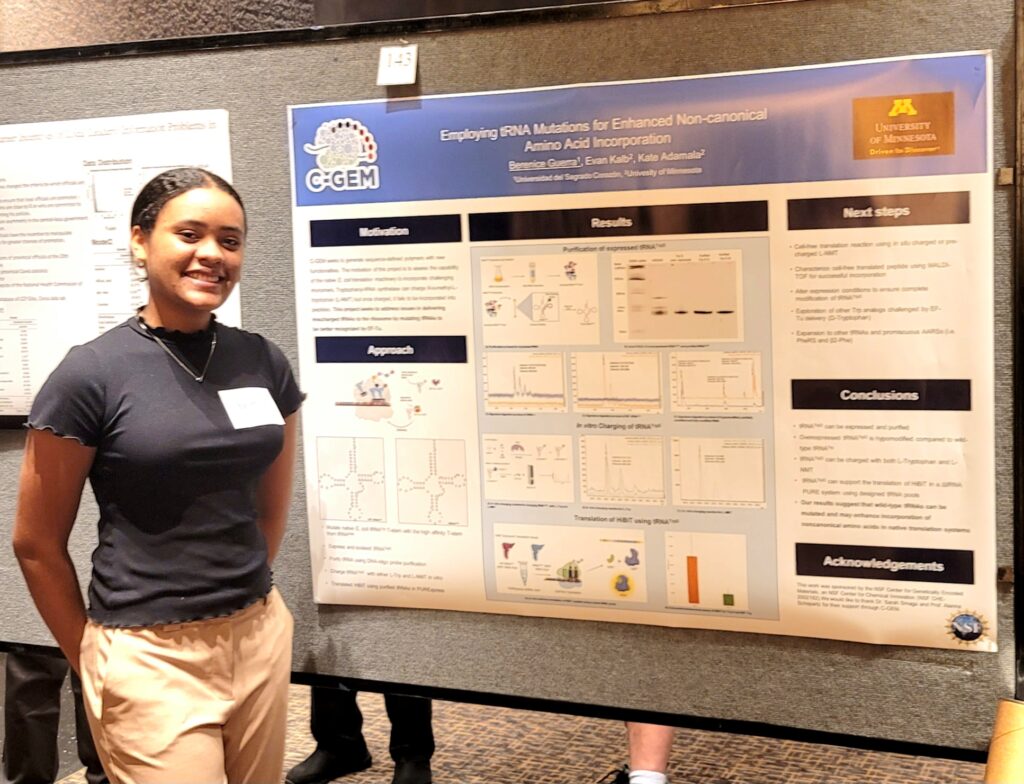
Summer Undergraduate Research Expo
Congratulations to Berenice Guerra and Hailee Aro for presenting their posters at the Summer Undergraduate Research Expo.
2023-07-09 22:21:32
new publication
|
|
Encyclopedia of Astrobiology; |
2023-06-07 15:30:57
new publication
|
|
Cell-Free Expression System Derived from a Near-Minimal Synthetic Bacterium; |
2023-05-29 20:16:48
SynCell poster prizes
Congratulations to Evan Kalb and Abbey Robinson for winning poster prizes at SynCell2023.

2023-05-14 21:26:30
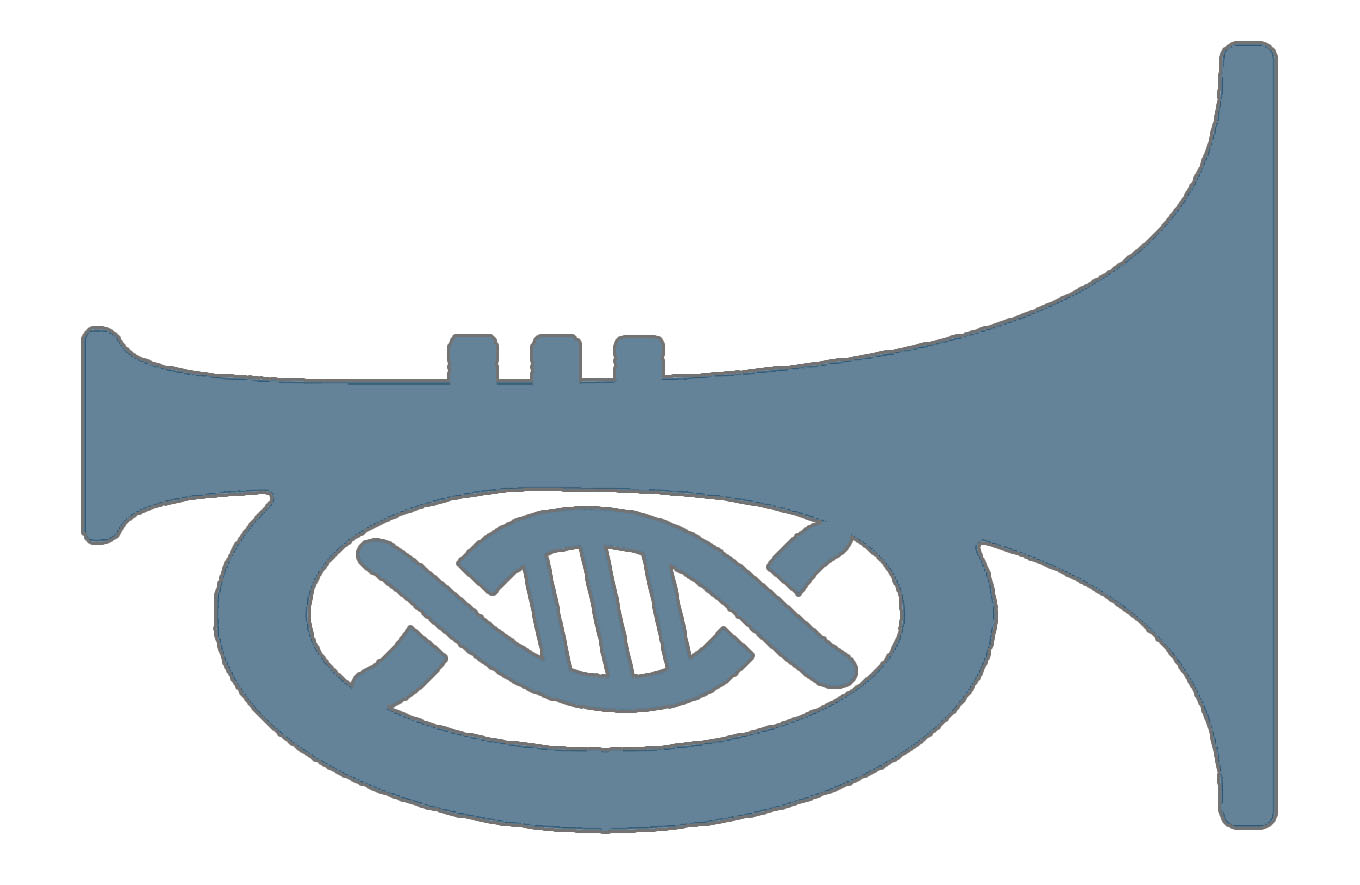
Trumpet in the news
Kate and Judee talk about Trumpet biocomputing platform and practical applications for medicine.

2023-05-04 21:23:22
Trumpet biocomputing
Our Trumpet platform summarized with emphasis on biomedical applications.

2023-04-26 20:06:05
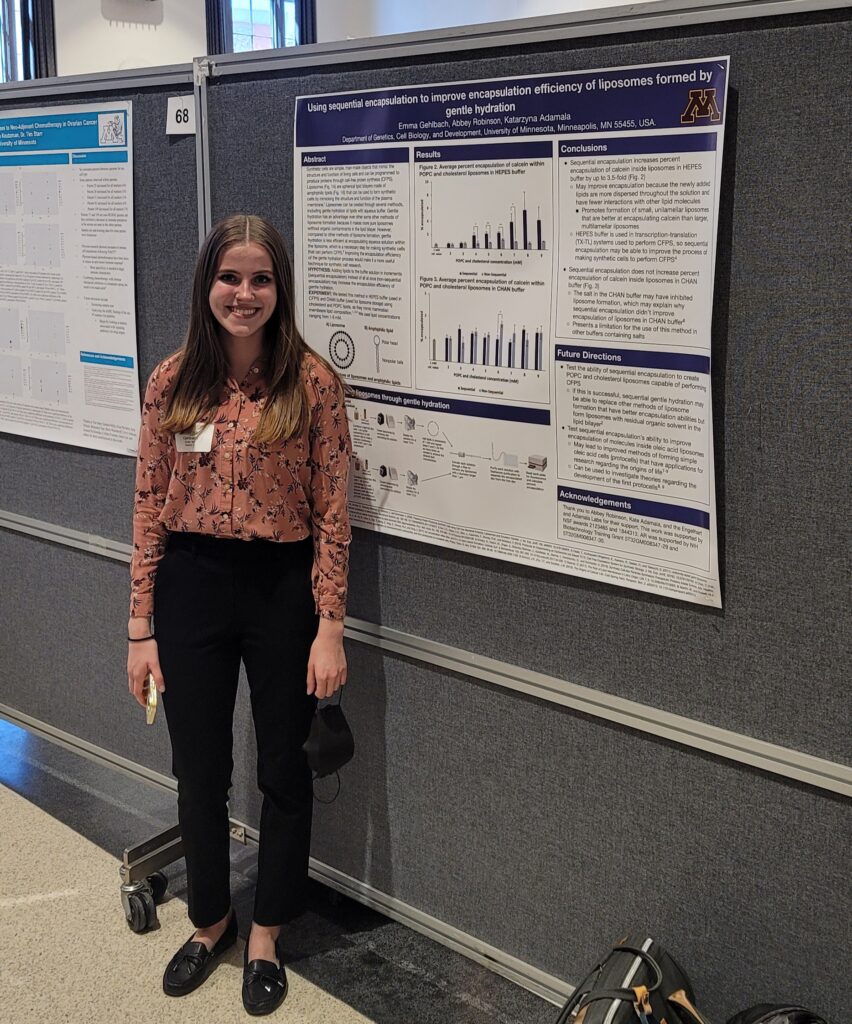
Emma's poster
Congratulations to Emma Gehlbach for presenting her poster at the Undergraduate Research Symposium.
2023-04-20 14:12:57
new publication
|
|
A gene expression control technology for cell-free systems and synthetic cells via targeted gene silencing and transfection; |
2023-04-10 19:10:42
Building life in the lab - Reason with Science
Kate talked about building life in the lab on the Reason with Science channel. Recording on youtube or podcast website.
2023-04-05 03:25:27
new publication
|
|
New Aequorea fluorescent proteins for cell‐free bioengineering; |
2023-03-06 23:45:31
Origins initiative
Kate participated in the AAAS panel announcement of the new Origins Initiative.
https://www.phy.cam.ac.uk/news/humanitys-quest-discover-origins-life-universe

2023-03-02 23:37:57
space panel
Kate participated in space exploration panel at the Swiss Embassy in DC, hosted by His Excellency Embassador Pitteloud.

2023-02-10 15:53:35
new publication
|
|
Parasites, infections and inoculation in synthetic minimal cells; |
2023-01-23 04:01:14
new publication
|
|
T7Max transcription system |
2023-01-02 18:46:36
Neha Anwar podcast on synthetic cells
Kate was guest of the Neha Anwar podcast, talking about biomedical and other practical applications of synthetic cell engineering.
Synthetic cell engineering podcast.
2022-12-28 02:13:47
YoloTalks: Creating artificial cells
Kate was guest of the YoloTalks podcast, talking about artificial cells and space exploration. Youtube: Creating artificial cells, and Spotify version.
2022-11-16 21:24:24
Life as we don't know it
Our lab was featured in CBS Connect article "Life as we don't know it".
2022-09-12 23:58:11
webinar Building a Synthetic Biology Platform for Drug Delivery
Kate participates in webinar "Building a Synthetic Biology Platform for Drug Delivery" organized by The Scientist.
![]()
2022-08-31 18:26:22
new publication
|
|
A ubiquitous amino acid source for prokaryotic and eukaryotic cell-free transcription-translation systems |
2022-08-15 17:09:42
new publication
|
|
Engineering Ribosomes to Alleviate Abiotic Stress in Plants; |
2022-08-09 22:28:42
Dr. Gaut!
The first student graduated from our lab. Congratulations, Dr. Gaut!

2022-07-28 15:26:27
Synthetic cell pathway to FDA
![]()
https://www.statnews.com/2022/07/26/fda-develop-framework-evaluate-synthetic-cells/
2022-05-25 23:28:45
SynCell 2022
Evan presented a poster, and Orion and Kate did a liposome demo at SynCell 2022. 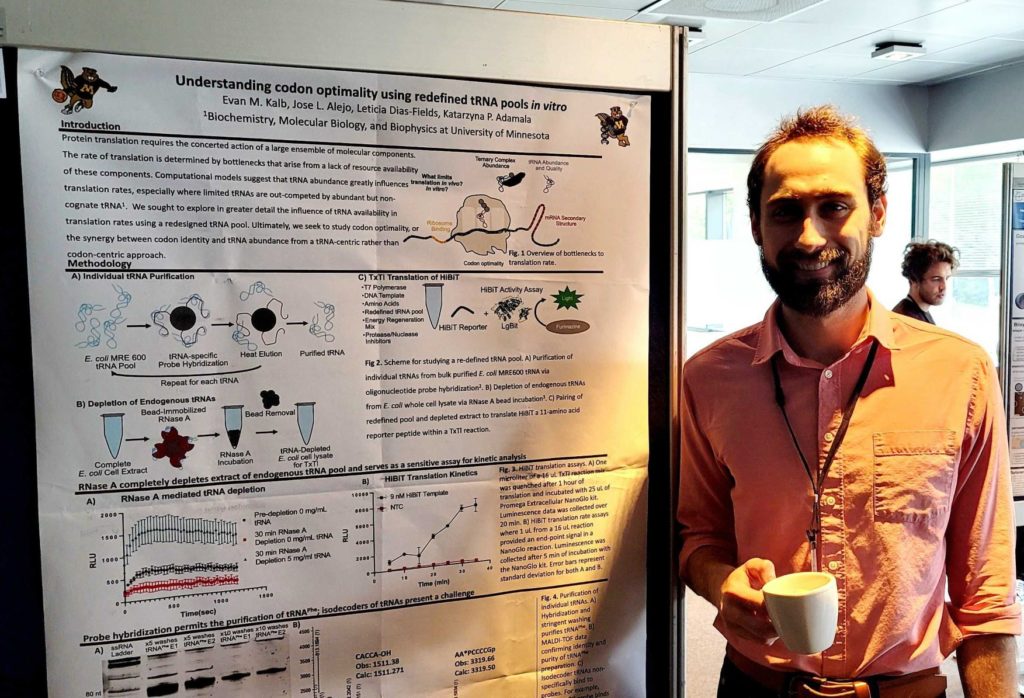

2022-05-11 02:39:00
new publication
|
|
Expanding luciferase reporter systems for cell-free protein expression; |
2022-05-08 18:39:44
Building Artificial Cells with Kate Adamala | Late Night Conference
Kate Adamala wants to make life from scratch! As Professor at University of Minnesota working on the origins of life and building a synthetic cell, Kate’s work touches on astrobiology, synthetic cell engineering, and biocomputing. Kate and her team create tiny bioreactors. These have applications in synthetic biology, drug development and biosensing. Building Artificial Cells with Kate Adamala | Late Night Conference with Wilhelm Huck 2x05.
2022-05-06 23:15:13
Fargo Theater public lecture
Kate gave talk at the Fargo Theater on engineering synthetic life.

2022-05-01 23:50:51
new publication
|
|
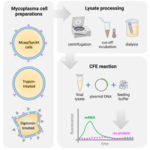
Traditional protocols and optimization methods lead to absent expression in mycoplasma cell-free gene expression platform |
2022-04-25 19:02:59
The life of our Last Universal Common Ancestor
Synthetic biology allows us to build minimal cell-like systems, to investigate origin of life and build tools for space exploration.
The life of our Last Universal Common Ancestor | LAS 2021.
2022-04-04 23:22:53
new publication
|
|
Akaby - cell-free protein expression system for linear templates |
2022-03-21 17:48:02
new publication
|
|
Diffusion control in biochemical specificity; |
2022-03-14 21:08:14
new publication
|
|
Switchable DNA-based Peroxidases Controlled by a Chaotropic Ion; |
2022-03-08 23:22:12
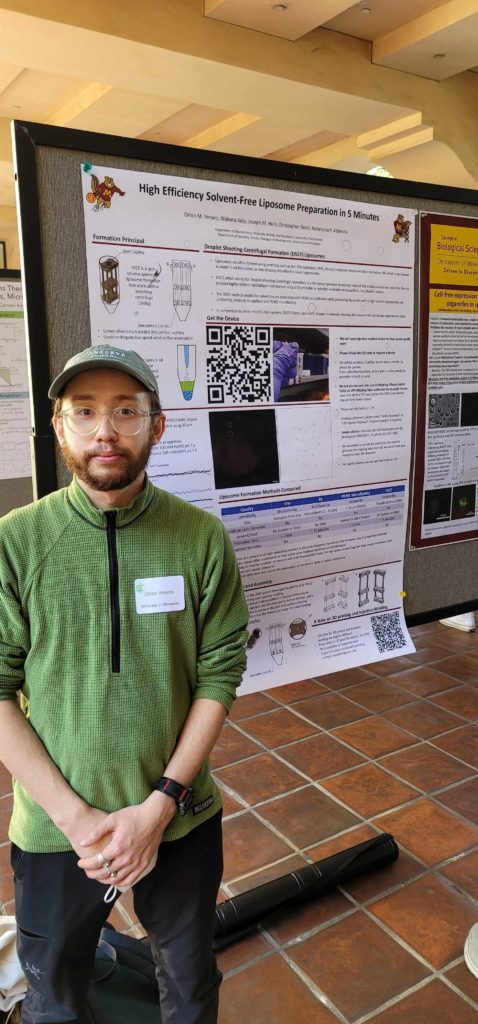
Build-a-Cell workshop
Abbey, Anders and Orion presented posters at Build-a-Cell Workshop 8 at Caltech.

2022-02-18 17:26:59
new publication
|
|
Making Security Viral: Shifting Engineering Biology Culture and Publishing; |
2022-01-29 19:15:24
Akaby
Akaby strain for linear TxTl is now available to all non-profit users: Akaby strain and information

2022-01-11 23:43:50
new publication
|
|
Liposome Preparation by 3D-Printed Microcapillary-Based Apparatus; |
2022-01-06 19:29:33
Artificial cells’ valuable niche in medicine
Advanced Science News piece:
Artificial cells with specialized internal chemistries could revolutionize how we approach precision medicine.

https://www.advancedsciencenews.com/artificial-cells-valuable-niche-in-medicine/
2021-12-29 00:37:48
new publication
Nathaniel J. Gaut, Jose Gomez-Garcia, Joseph M. Heili, Brock Cash, Qiyuan Han, Aaron E. Engelhart, Katarzyna P. Adamala
ACS Syn Bio, 2022, https://doi.org/10.1021/acssynbio.1c00519
webinar on synthetic cell engineering
Kate participates in The Dissenter webinar "Synthetic Cells, Cell Evolution, the Origin of Life, and Astrobiology".
2021-11-18 19:42:25
Engineering synthetic cells for medicine
Build-a-Cell video: Engineering synthetic cells for medicine
2021-11-09 02:23:48
EBRC podcast on safety of synthetic cells
Kate was a guest of EBRC In Translation podcast, discussing science, safety and security of synthetic cell engineering ebrcintranslation.buzzsprout.com/1581817/9465178-9-building-cells-and-synbio-in-space-w-kate-adamala

2021-11-02 04:32:57
new publication
|
|
Build-a-Cell: Engineering a Synthetic Cell Community |
2021-11-02 04:00:26
new publication
|
|
Synthetic Cells in Biomedical Applications |
2021-10-29 19:18:44
Liposome formation device
Our simple, 3D printed liposome formation device is now available to all non-profit users: device, protocols and tutorials
2021-10-04 23:30:52
Synthetic cells for drug delivery
CRS News published our text about synthetic cells in drug delivery applications.
Synthetic cells: happy middle between liposome drug delivery and engineered natural cells

2021-10-04 17:33:41
new publication
|
|
Building a community to engineer synthetic cells and organelles from the bottom-up |
2021-10-04 15:28:32
new publication
|
|
Toward synthetic life: Biomimetic synthetic cell communication |
2021-09-29 19:12:54
Trumpet platform
TRUMPET: Transcriptional RNA Universal Multi-Purpose GatE PlaTform is our new biocomputing tool. The web based logic gate design tool is now live on trumpet.bio

2021-08-04 23:37:21
iBiology synthetic cells
Kate's iBiology talks on synthetic cells have been published.
Synthetic Cells: Building Life to Understand It

Part 1: Synthetic Cells: Building Life to Understand It

Part 2: Synthetic Cells: Approach and Applications
2021-03-25 19:05:38
webinar Synthetic Cells · Artificial Life · Brain-Computer Interfaces · Space Exploration
Kate participates in webinar Synthetic Cells · Artificial Life · Brain-Computer Interfaces · Space Exploration hosted by the Molecular Programming Interest Group.

2021-02-10 16:07:52
new publication
|
|
Reconstituting Natural Cell Elements in Synthetic Cells |
2021-02-05 16:23:47
Santa Fe Institute
Kate participated in the Santa Fe Institute workshop Origins of Life: the Possible and the Actual.
2021-01-22 19:00:10
Life but not alive - Origins Center
Webinars of the Origins Center, the Netherlands: "Life but not alive"
Life but not alive.
2020-12-12 22:36:53
new publication
|
|
Spatial multiplexing of fluorescent reporters for dynamic imaging of signal transduction networks |
2020-12-09 05:41:59
Space Explr podcast on building synthetic cells
Kate was a guest of Space Explr podcast, discussing Science And Engineering of Building Synthetic Cells https://www.youtube.com/watch?v=4fznl9wLtmw
2020-12-05 23:20:31
The Darwin
Kate was guest speaker at Darwin 2020, One of India's Biggest Evolutionary Movements in Biology

2020-11-25 05:20:14
Andromeda Strain and Meaning of Life
Kate participated in Santa Fe Institute podcasts on the Andromeda Strain and Meaning of Life.
with Sara Walker:
https://www.youtube.com/watch?v=TH7O5IQpC1A
with Chris Kempes
https://www.youtube.com/watch?v=79qzllZrroc
2020-10-29 18:53:09
Probable Meets Possible: When Life Gets Weird
Kate Adamala and Trinity Hamilton from the College of Biological Sciences at the University of Minnesota discuss how tiny life forms on the fringes may illuminate huge truths about life in the universe. What do microbes that live on glaciers and in snow on Earth tell us about the potential for life on Mars? What can a cell made from scratch in a lab reveal about how life may have originated here on Earth—and beyond? This research on extremophiles and synthetic cells provides a unique take on life that may shed important insights into space travel and extraterrestrial life.
Probable Meets Possible: When Life Gets Weird.
2020-10-23 05:38:59
Probable Meets Possible: When Life Gets Weird
Popular science podcast on extremes of life: Probable Meets Possible: When Life Gets Weird, by CBS and Bell Museum:
https://www.youtube.com/watch?v=82MmOBJK9B0
2020-09-22 20:48:17
Community of Biotechnology talk
Kate gave a seminar for the Community of Biotechnology.
https://www.facebook.com/cbiotechnology/videos/680207569301004/
2020-09-21 23:49:47
new publication
|
|
Rapid deployment of smartphone‐based augmented reality tools for field and online education in structural biology |
2020-06-30 18:58:01
Conversations with Aanika E10: Creating Synthetic Cells with Dr. Kate Adamala
Aanika Biosciences presents conversation series on topics of cutting edge synthetic biology.
Conversations with Aanika E10: Creating Synthetic Cells with Dr. Kate Adamala.
2020-06-19 05:34:50
Conversations with Aanika E10: Creating Synthetic Cells
Short interview about synthetic cell engineering: https://www.youtube.com/watch?v=kDAPfVLkTts
2020-06-05 01:33:56
new publication
|
|
Highly specific, multiplexed isothermal pathogen detection with fluorescent aptamer readout |
2020-06-01 00:09:42
Quanta podcast
Kate helped with Quanta Magazine Bacterial Complexity Revises Ideas About ‘Which Came First?’ podcast.
https://www.quantamagazine.org/bacterial-organelles-revise-ideas-about-which-came-first-20190612/
2020-05-25 19:49:01
new publication
|
|
Toward artificial photosynthesis Nathaniel J Gaut and Katarzyna P Adamala Science Vol. 368, Issue 6491, pp. 587-588 journal link |
2020-04-09 18:47:27
Lab Work Under Isolation
Opinion on lab operations in The Scientist
https://www.the-scientist.com/opinion-lab-work-under-isolation-67398
2020-04-08 18:49:26
EBRC Virtual Annual Meeting
Kate chaired the first session of the EBRC virtual annual meeting:
Cell-Free session.
2020-03-27 05:13:47
Center for Genome Engineering
Kate gave seminar for the Center for Genome Engineering, in the new era of zoomeetings.

2020-03-05 05:42:47
Synthetic cell talk online
Kate's Waterloo seminar is posted online. Primer on synthetic cell work in our lab and little bit of the overview of the field.
2020-02-07 05:03:55
Waterloo Institute of Complexity and Innovation
Kate gave a seminar at the Waterloo Institute of Complexity and Innovation.
2020-01-31 05:01:50
Build-a-Cell workshop #6
Theodore, Orion, Kaitlin, Wakana and Kate went to Build-a-Cell Workshop #6 at NASA Ames.
2020-01-24 04:58:24
Breakthrough Foundation Panspermia Workshop
Kate is at Breakthrough Foundation Panspermia Workshop at the ASU Beyond Center for Fundamental Concepts in Science.
2019-12-04 01:08:00
Cell-Free Systems Boston
Kate, Judee and Lauren are at Cell-Free Systems Conference in Boston.
2019-11-21 02:17:36
P3 Dystopia
Kate's episode on P3 Dystopia show is now live.
https://sverigesradio.se/artikel/7345117
2019-11-20 01:03:27
Iowa State seminar
Kate was visiting Department of Genetics, Cell Biology and Development at the Iowa State University.
https://www.gdcb.iastate.edu/gdcb-seminar-%E2%80%94-bioengineering-synthetic-cells
2019-11-18 05:16:06
new publication
|
|
Connectome of synthetic cells: Comment on “What would a synthetic connectome look like?” by Ithai Rabinowitch; |
2019-11-14 00:49:07
new publication
|
|
Enabling community-based metrology for wood-degrading fungi; |
2019-11-12 01:16:30
iGem 2019

Kate participated in 2019 iGem Jamboree as a judge and as an adviser to the amazing Brown - Stanford - Princeton iGem team, supporting the Astropharmacy project.
https://2019.igem.org/Team:BrownStanfordPrinctn/Team
SynCell EU
Kate represented Build-a-Cell community at the SynCell EU meeting in Madrid.

2019-10-16 01:32:19
NSTC Interagency Synthetic Biology Workshop
Kate represented Build-a-Cell at the NSTC Interagency Synthetic Biology Workshop
https://ebrc.org/nstc-interagency-synthetic-biology-workshop/
2019-10-06 00:59:01
new publication
Tanner G. Hoog, Matthew R. Pawlak, Lauren M. Aufdembrink, Benjamin R. Bachan, Matthew B. Galles, Nicholas B. Bense, Katarzyna P. Adamala, Aaron E. Engelhart
https://doi.org/10.1101/784561
EBRC Global Forum For Engineering Biology: Initial Review Of Synthetic Biology National Strategies
Kate represented Build-a-Cell community at the EBRC Global Forum For Engineering Biology: Initial Review Of Synthetic Biology National Strategies
https://ebrc.org/https-ebrc-org-global-forum-for-engineering-biology/
2019-08-29 19:20:47
Liposome calculation tool
We published interactive spreadsheet for liposome math: volyume, size, encapsulation rate and liposome numbers.

2019-08-16 01:38:48
Build-a-Cell Workshop #5
Kate, Judee and Wakana participated in Build-a-Cell Workshop #6 in Boston.
http://buildacell.io/engineering/workshop5/

2019-08-09 01:43:44
DNA25
Kate gave talk at DNA25 in Seattle.
http://misl.cs.washington.edu/events/dna25/
2019-08-05 18:55:29
Origins of Life: Chemical Comminalties - Early Metabolisms - An Introduction
ComplexityExplorer.org course 'Origins of Life
Origins of Life: Chemical Comminalties - Early Metabolisms - An Introduction by Kate Adamala.
2019-07-31 01:50:53
Designer Biology
Kate gave talk at the Designer Biology: From proteins and cells to scaffolds & materials; in Newcastle UK
http://designer-biology.org/
![]()
2019-07-30 02:10:49
Mindscape podcast
Kate's episode on Sean Carroll's Mindscape podcast is live.
https://www.preposterousuniverse.com/podcast/2019/07/22/56-kate-adamala-on-creating-synthetic-life/

2019-07-25 01:45:17
SMBE
Kate gave talk at the Society for Molecular Biology and Evolution annual meeting in Manchester, UK
2019-07-19 18:09:40
new publication
|
|
Biology on sample size of more than one |
2019-07-04 02:20:53
SEED 2019 - Build-a-Cell
Interview with Kate about Build-a-Cell
2019-06-28 02:26:25
Interplanetary Festival
Kate participated in origin of life panel at the Interplanetary Festival in Santa Fe.
https://interplanetaryfest.org/

2019-06-25 02:21:57
SEED 2019
Kate organized Build-a-Cell session at SEED 2019.
![]()
2019-05-29 18:46:20
Life, But Not Alive: Breakthrough Discuss 2019
Breakthrough Discuss 2019 was held at UC Berkeley April 11-12, 2019. The theme, “Migration of Life in the Universe,” brought together an all-star group of astronomers, biologists, chemists and other experts to discuss whether and how primitive life can move between planets and stars, and if that is how life began on Earth.
Kate Adamala | Life, But Not Alive: Breakthrough Discuss 2019 .
2019-04-30 17:28:22
OoLALA seminar
Kate participated in OoLALA seminar series, talking about minimal life.

2019-04-17 17:25:35
FNANO 2019
Kate gave seminar at Foundations Of Nanoscience: Self-Assembled Architectures And Devices (Fnano19).
2019-04-12 17:21:29
Breakthrough Discuss
Kate, Aaron, Joe, Tanner, Judee and Wakana participated in 2019 Breakthrough Discuss meeting.
2019-03-29 22:55:02
New publication
Sivan Kaminski Strauss, (...) Katarzyna P. Adamala, (...), Orna Dahan, Yitzhak Pilpel;
Plos Biology, doi.org/10.1371/journal.pbio.3000182;
local copy
publisher website link
Build-a-Cell Workshop #4
Kate, Nathan and Chris participated in Build-a-Cell Workshop #4.

2018-12-17 16:58:15
The Leap to Life
Our lab is proud to be a part of new Templeton Foundation team to study chemical emergence of life.
Exploring the informational transitions bridging simple chemistry and minimal life
2018-11-30 02:31:12
SeMiSynBio kick-off meeting
Kate presents at the kick-off meeting for the SemiSynBio project, talking about biocomputing using synthetic cells.
2018-11-23 02:17:13
Artificial cells gain communication skills
Great editorial in Science intorducing new, exciting paper and highlighting interesting aspects of building synthetic cells.
2018-11-16 02:24:18
ASU-SFI Theory in Origins of Life Working Group
Kate participated in the ASU-SFI Theory in Origins of Life Working Group in Phoenix.
2018-11-08 01:50:43
science fair
Nathan, Lauren, Joe, Matt and Adam brough in vitro transcription demo to science fair at Andersen United Community School.

2018-11-07 02:09:19
How biologists are creating life-like cells from scratch
Excellen editorial in Nature, describing rationale of engineering synthetic life, and the most common approaches to various problems we're facing en route to a ynthetic cell. 
Local copy pdf
2018-10-30 02:20:11
Exploring life's origins
Kate went to Santa Fe Institute Workshop "Major Transitions in Life: Origins to Translations", a kick-off meeting for Exploring life's origins project at Santa Fe Institute.
2018-10-14 04:15:32
new publication
Lin Jin, Aaron E. Engelhart, Weicheng Zhang, Katarzyna Adamala, Jack W. Szostak
J. Am. Chem. Soc. 2018, 140, 44, 15016-15021, doi.org/10.1021/jacs.8b09617
Synthetic Biology for Defense Workshop
Kate presented at Synthetic Biology for Defense Workshop in Arlington, Virginia.
2018-09-17 01:53:21
Jose won HHMI fellowship
Congratulations for Jose Alejo, he was named this year's Hanna H. Gray Fellow by the Howard Hughes Medical Institute.
Hanna Gray Fellows

2018-08-30 01:59:56
BaSyC symposium
Kate presented at 1st International Symposium on Building a Synthetic Cell , amazing meeting dedicated to construction of synthetic cells.


2018-08-29 01:09:09
New publication
Joseph M. Heili, Jose Gomez-Garcia, Nathaniel J. Gaut, Brock W. Cash, Lauren M. Aufdembrink, Brent A. Heffron, Joshua D. Shirley, Erin E. Carlson, Katarzyna P. Adamala, and Aaron E. Engelhart
J. Chem. Educ.,, 2018, 95 (10), pp 1867–1871; DOI: 10.1021/acs.jchemed.7b00735
local copy pdf publisher website
Build-A-Cell workshop #3
We hosted Build-a-Cell Workshop #3 in Minneapolis.
Amazing group of people discussing ways to make life.


2018-07-22 02:10:34
The Next Revolution: Genome Engineering 2018
Kate presented at Annual Conference of the Genome Writers Guild

2018-07-17 02:25:13
Biocomputing
Our lab is part of a new project to support large scale developement of biocomputing technologies, jointly funded by the National Science Foundation (NSF) and the Semiconductor Research Corporation (SRC).

2018-06-21 01:45:39
synthetic cells in space!
Our synthetic cell experiment launched today at NASA Wallops!
The take off was luckily uneventfull, we're excited to start analyzing data now.


2018-05-23 02:07:06
Three Minute Thesis
Congratulations to Nathan Gaut for winning second place in the UMn 3MT finals!

2018-05-14 02:07:34
NSF Synthetic and Artificial Cells Workshop
Kate attended NSF Synthetic and Artificial Cells Workshop in Alexandria VA.
2018-05-11 02:10:34
Synthetic Biology USA Congress
Kate gave talk on bioengineering with synthetic cell at 3rd Synthetic Biology USA Congress
2018-02-07 16:48:12
New publication

Build-a-Cell Workshop #2
Kate and Brock were attending Build-a-Cell #2 Workshop, hosted at Stanford.
buildacell.io

2017-12-23 17:15:20
What is life podcast
Kate participated in What is life conversation about origins and earliest evolution of life , led by Carl Zimmer at Caveat in New York City.

2017-12-01 20:10:24
Princeton Envision
Kate talked and run a workshop at 2017 Envision meeting at Princeton.

2017-11-09 20:05:56
iGem 2017
Kate is judging 2017 iGem during 2017 Giant Jamboree in Boston.

2017-10-09 20:21:47
NASA Biomimicry
Kate talked about synthetic cell technologies for biomimicry at Nature-Inspired Exploration for Aerospace Summit at Ohio Aerospace Institute.

2017-10-06 20:18:50
Cell mediated therapy
Kate visited Abbvie to talk about using synthetic cells for cell-mediate therapy.
2017-09-04 15:42:26
EON Workshop on Universal Biology
Kate was a speaker at EON Workshop on Universal Biology at ELSI in Tokyo.

2017-08-06 22:06:09
Fab13
Kate was a speaker at Fab13 Symposium during Fab13 meeting in Santiago, Chile.


2017-07-30 02:20:28
From chemicals to life: Scientists try to build cells from scratch
Interesting summary of the origins of Build-a-Cell initiative.
2017-07-20 23:37:25
ISSOL 2017
Kate gave a talk, and various lab members had names on few posters during XVIIIth International Conference on the Origin of Life

2017-07-13 23:36:35
Genetic circuits in synthetic cells
Kate published commentary on using synthetic cells to prototype cell circuits in Cell Systems journal.
2017-07-10 17:30:10
Personalized medicine
Kate's Medium post Why should we personalize medicine?
2017-06-30 22:45:00
new publication
|
|
What Is the Role of Circuit Design in the Advancement of Synthetic Biology? |
2017-06-16 04:51:21
SB7
Kate is giving a talk, and Nathan is presenting a poster at SB7, The Seventh International Meeting on Synthetic Biology in Singapore.

2017-06-11 04:58:05
Visit to Tsinghua University
Kate and Aaron are visiting Ting Zhu at Tsinghua University in Beijing.
2017-06-09 05:04:29
New Lab member
Welcome Brock Cash to the lab!
2017-04-26 05:22:14
Brains on! podcast
Aaron and Kate did NPR podcast on the origin of life - Brains on!

2017-04-20 16:40:42
Innovations in Biology - Advancing Biomanufacturing
Kate gave a talk, while Joe and Nathan presented posters at Grand Challenges Vision Symposium Innovations in Biology - Advancing Biomanufacturing

2017-04-19 12:01:24
Congratulations Jose!
Jose Gomez-Garcia won 2nd place poster presentation prize at CBS Postdoc Symposium.

2017-04-09 01:05:29
New publication
New Lab member
Welcome Kei Takahashi to the lab!
2017-02-01 15:54:33
Biological Complexity: Can it be Quantified?
Kate at Physics of Living Matter workshop at the Beyond Center, ASU.
2017-01-30 15:59:06
@UCSF
Kate at UCSF, seminar, "Life but not alive: building a cell from scratch"
2017-01-17 15:59:57
Ask Me Anything about synthetic cells
Reddit AMA on synthetic minimal cells, answering questions from the public about our work. AskScience AMA Synthetic Cells.

2017-01-06 16:02:39
TEDx Talk on Synthetic life
Kate presented concept of building synthetic minimal cells, with its biotechnological, biomedical and basic science implications, in a TEDx talk "Life but not alive".
2016-12-20 16:09:29
New lab member
Welcome Joe Heili to the lab!
2016-11-14 02:03:42
New publication
Engineering genetic circuit interactions within and between synthetic minimal cells;
Katarzyna P. Adamala*, Daniel A. Martin-Alarcon*, Katriona R. Guthrie-Honea, Edward S. Boyden;
Nature Chemistry, 9, 431-439, 2017, doi:10.1038/nchem.2644; *equal contribution
publisher website link
local copy pdf
Masonic Cancer Center
Our lab become affiliated with the Masonic Canter Center.
We're proud to become part of this amazing research community.
2016-10-24 16:11:38
Synthetic cells in the news
So we are working on building "simplified cells" now. Associated Press noticed (local copy pdf).
So did MPR news (local copy pdf).
2016-10-15 16:13:35
New Lab member
Welcome Jose Gomez-Garcia to the lab!
2016-10-01 23:13:31
New publication
Conference
Kate at BioHack the Planet!

2016-09-19 03:26:44
New publication

Astrobiology Primer 2.0.;
Domagal-Goldman and Wright et al. (includes chapter editor K.P.Adamala)
Astrobiology, 2016, 16(8):561-653. doi:10.1089/ast.2015.1460
publisher website link
RNA supergroup
Our lab become member of the University of Minnesota RNA Supergroup.
2016-08-14 16:15:50
HTGAA
How to grow almost Anything?
HTGAA Class is back in 2016. More students, more sites.
2016-08-09 21:10:41
Spatial multiplexing for multisignal cellular imaging
G Xu, E Boyden, KD Piatkevich, K Adamala
US Patent App. 15/099,232, 2016
outside link
2016-08-09 21:01:32
Pumilio Domain-based Modular Protein Architecture for RNA Binding
ES Boyden, KP Adamala, DA Martin-Alarcon
US Patent App. 14/995,169
outside link
2016-08-01 16:18:43
FAB 2.0
The theme of the 12th international Fab Lab conference in Shenzhen, China, is fab labs that make fab labs: systems that makes systems, applying the biology concept of self-replicating systems into the fabrication world.

Kate is giving a talk on synthetic life during the plenary Symposium.
What if we build life instead of growing?
Or what if we grow machines instead of building?
2016-05-02 03:29:12
New Publication
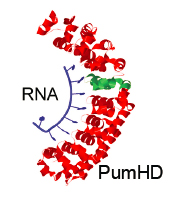
Programmable RNA-binding protein composed of repeats of a single modular unit;
Katarzyna P. Adamala*, Daniel A. Martin-Alarcon*, and Edward S. Boyden;
PNAS, 2016, 10.1073/pnas.1519368113; *equal contribution
local copy pdf
publisher website link
TEDx talk
Kate speaks at TEDxEsteeLauderCompanies event at MoMA in New York City.
2016-03-21 03:30:50
New publication
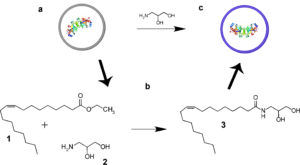
Collaboration between primitive cell membranes and soluble catalysts;
K. Adamala*, A. E. Engelhart* and J. W. Szostak;
Nature Communications, 2016, doi:10.1038/ncomms11041; *equal contribution
local copy pdf
publisher website link
Floating FAB LAB Amazon
The FabLab Lima and Konrad Adenauer Foundation organised workshop Floating FAB LAB Amazon.
We discussed the opportunities for using synthetic minimal cells as biosensors in the Floating FabLab on the Amazon river.

2015-09-06 16:22:36
Fab Academy meets synthetic biology.
Introducing How To Grow (Almost) Anything
The first synth bio FabLab Academy includes Kate's unit on synthetic minimal cells.
2015-07-09 03:43:12
New publication
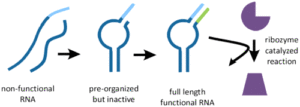
Generation of Functional RNAs from Inactive Oligonucleotide Complexes by Non-enzymatic Primer Extension;
K. Adamala*, A. E. Engelhart* and J. W. Szostak;
J. Am. Chem. Soc., 2015, 137 (1), pp 483 - 489, DOI: 10.1021/ja511564d; *equal contribution
local copy pdf
publisher website link
New publication
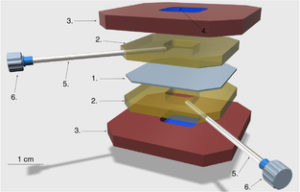
Construction of a Liposome Dialyzer for preparation of high-value, small-volume liposome formulations;
K. Adamala*, A. E. Engelhart*, N. Kamat, L. Jin and J. W. Szostak;
Nature Protocols, 2015, 10(6), pp 927-938; *equal contribution
local copy pdf
publisher website link
New publication
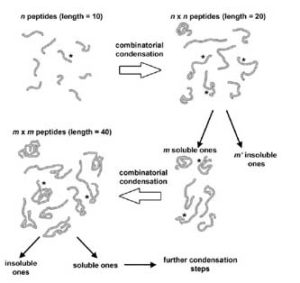
Open questions in origin of life: experimental studies on the origin of nucleic acids and proteins with specific and functional sequences by a chemical synthetic biology approach;
Adamala K, Anella F, Wieczorek R, Stano P, Chiarabelli C, Luisi PL;
Comput Struct Biotechnol J. 2014;9:e201402004
local copy pdf
publisher website link
New publication

Non-enzymatic template-directed RNA synthesis inside model protocells;
K. Adamala and J.W. Szostak,
Science 342 (2013) 1098 - 1100;
local copy pdf
publisher website link